Tutorials
MADS
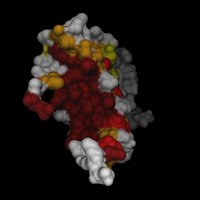
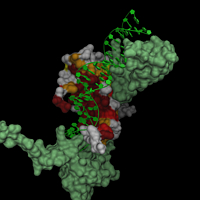
This is from the PDB file 1MNM (yeast matalpha2/mcm1/dna ternary transcription complex, S. Tan, T.J. Richmond, Nature 391 1998).
I did a quick search through the database to find similar sequences to MCM1 (containing the MADS box), aligned them and analyzed the result with Alscript (G.J. Barton, Protein Engineering 6). I used the similarity codes 4,6,8 and 10 to select residues in the structure with increasing conservation among the sequences.
Download this tutorial: mads.tar.gz
// from 1MNM (Tan et. al. (1998) Nature 391) // set variables set c10 (rnum=18,26,35,38,39,42,45,46,47,49,50) set c8 (rnum=20,23,32,36,40,43,54,58,60,61,72) set c6 (rnum=21,28,31,34,37,44,51,52,56,62,64,67,69) set c4 (rnum=25,55,57,59,63,70,71) // load datasets load mads.pdb load mads_p1 -type msms -name smcm1 load mads_p2a -type msms -name smat2a load mads_p2b -type msms -name smat2b load mads_d -type msms -name sdna // center scene center [.mads] // generate trace objects .mads new -type trace -name mcm1 -sel chain=A,B .mads new -type trace -name mat2 -sel chain=C,D .mads new -type trace -name dna -sel chain=E,F // attach surfaces to corresponding parts in .mads .mads restrict chain=A,B .smcm1 attach .mads .mads restrict chain=C .smat2a attach .mads .mads restrict chain=D .smat2b attach .mads .mads restrict chain=E,F .sdna attach .mads .mads restrict // generate new surfaces .smcm1 new -name all .smat2 new -name all .sdna new -name all .smat2a new -name all .smat2b new -name all // color .smat2?.all set color=lightgreen .smat2?.all hide .sdna.all hide .smcm1.all set color=yellow -sel $c4 .smcm1.all set color=orange -sel $c6 .smcm1.all set color=red -sel $c8 .smcm1.all set color=red4 -sel $c10 .sdna.all set color=grey50 .sdna.all render t=0.4 .mads.dna render hsc scene autoslab |