Tutorials
Surface Dataset
A short introduction into displaying molecular surfaces.
Following the DINO philosophie, a molecular surface is not created by the program, but is generated externally with e.g. MSMS or MSP.
With MSMS, the PDB file is first converted into an .xyzr file, which contains the coordinates and the radius of each atom. (This is done with pdb_to_xyzr from the MSMS distro or with pdb2xyzr , a perl script which carries the definition file within and is slightly faster). This .xyzr file is then used as input for MSMS and the result are two files: a .vert and a .face file.
> pdb2xyzr NAME.pdb > NAME.xyzr > msms -if NAME.xyzr -of NAME > ls NAME.pdb NAME.xyzr NAME.face NAME.vert
Download this tutorial: surf.tar.gz
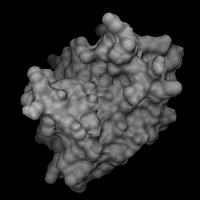
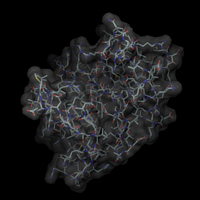
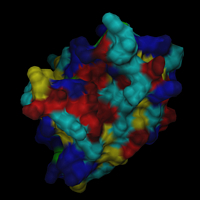
// load the structure dataset load iib.pdb -name iib // load the surface dataset load iib -type msms -name surf // attach the surface to the structure // (thereby assigning each surface point // to an atom) .surf attach .iib // center on the surface scene center [.surf] // create a surface object .surf new -name all // adjust the clipping planes scene autoslab // color the surface object according // to some underlying residue properties // the variables used here are predefined // by DINO, use 'var' to look at them .surf.all set color=blue -sel $basic .surf.all set color=blue4 -sel $basic2 .surf.all set color=red -sel $acidic .surf.all set color=red4 -sel $acidic2 .surf.all set color=cyan -sel $polar .surf.all set color=green -sel $aromatic .surf.all set color=yellow -sel $aliphatic // reset the color to white and make the surface // transparent to show the underlying structure .surf.all set color=white .iib new -name all .iib.all render custom .surf.all render t=0.4 // generate a solid surface // reset transparency and hide struct obj .surf.all render t=1 .iib.all hide scene set near+=23 .surf.all render solid,solidc=grey |